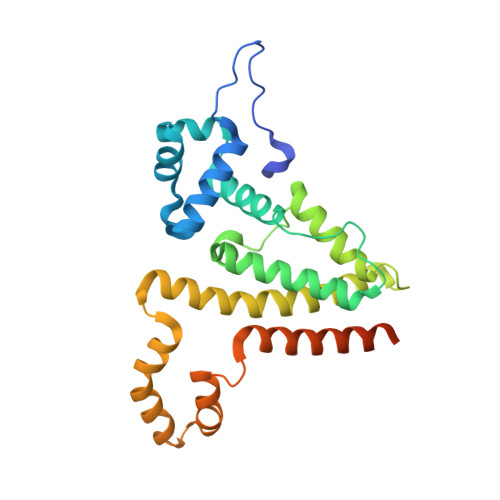
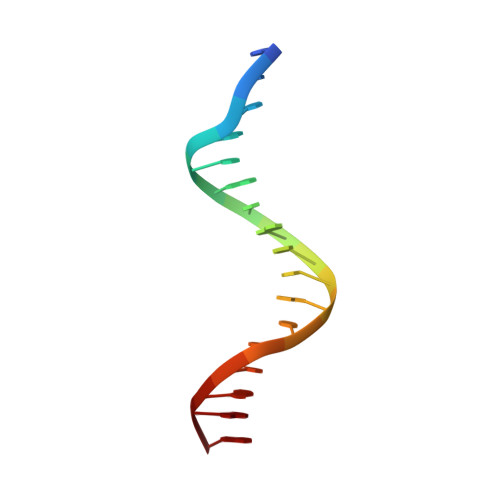
The Crystal Structure of the Tetr Family Transcriptional Repressor Simr Bound to DNA and the Role of a Flexible N-Terminal Extension in Minor Groove Binding.
Le, T.B.K., Schumacher, M.A., Lawson, D.M., Brennan, R.G., Buttner, M.J.(2011) Nucleic Acids Res 39: 9433
- PubMed: 21835774
- DOI: https://doi.org/10.1093/nar/gkr640
- Primary Citation of Related Structures:
3ZQL - PubMed Abstract:
SimR, a TetR-family transcriptional regulator (TFR), controls the export of simocyclinone, a potent DNA gyrase inhibitor made by Streptomyces antibioticus. Simocyclinone is exported by a specific efflux pump, SimX and the transcription of simX is repressed by SimR, which binds to two operators in the simR-simX intergenic region. The DNA-binding domain of SimR has a classical helix-turn-helix motif, but it also carries an arginine-rich N-terminal extension. Previous structural studies showed that the N-terminal extension is disordered in the absence of DNA. Here, we show that the N-terminal extension is sensitive to protease cleavage, but becomes protease resistant upon binding DNA. We demonstrate by deletion analysis that the extension contributes to DNA binding, and describe the crystal structure of SimR bound to its operator sequence, revealing that the N-terminal extension binds in the minor groove. In addition, SimR makes a number of sequence-specific contacts to the major groove via its helix-turn-helix motif. Bioinformatic analysis shows that an N-terminal extension rich in positively charged residues is a feature of the majority of TFRs. Comparison of the SimR-DNA and SimR-simocyclinone complexes reveals that the conformational changes associated with ligand-mediated derepression result primarily from rigid-body rotation of the subunits about the dimer interface.
Organizational Affiliation:
Department of Molecular Microbiology, John Innes Centre, Norwich Research Park, Norwich NR4 7UH, UK. tung.le@jic.ac.uk